Target enrichment
The genomes of insects and pathogens can be complex and difficult to decode. In order to facilitate the analyses and interpretation of the genomics of the forest enemies targeted by the BioSAFE project, we are developing a target enrichment approach that specifically targets and sequence regions of the genome that are useful to address questions that are relevant to the identification of their species or subspecies, their origin and traits. The BioSAFE platform uses a highly multiplexed amplification-based approach that is appropriate for handling environmental samples of unknown origin and various state of conservation. Next Generation Sequencing is used to identify specific changes in DNA by sequencing multiple gene targets within multiple samples in a single reaction. This approach is powerful, accurate and amenable to high throughput processing and it is widely used for cancer detection in the medical field. Our application of this approach for genomic biosurveillance of forest enemies will provide a powerful set of tools for detection and monitoring of invasive alien species.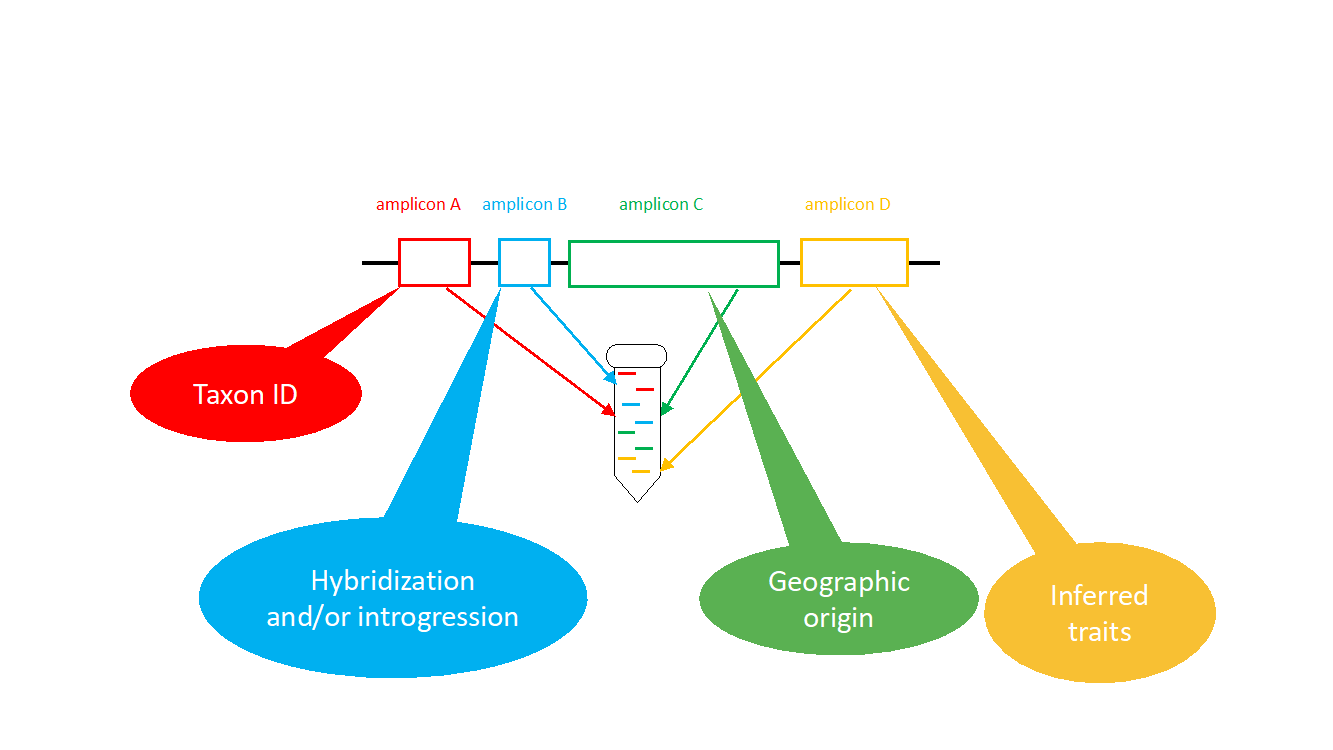
Figure 1. Example of an Ampliseq target enrichment panel that aims at sequencing multiple genome regions to address different questions.
How the technology will be used in this project?
Our aim is to design AmpliSeq panels (ThermoFisher) with multiple layers of amplicons (short fragments of DNA) that target different genomic regions of interest. For example, a panel can contain genome regions that are conserved across multiple taxa and will be taxonomically informative and others that we identified as being frequently involved in adaptive hybridization and introgression among taxa (Figure 1). Another layer contains amplicons that are highly polymorphic within species and are informative with regard to geographic origin or source and pathways of spread. A final layer contains genome regions that are associated with traits that are considered important for invasiveness such as flight capacity, cold tolerance, pathogenicity and host range (Figure 1).
The BioSAFE toolbox
A panel will be designed for each of the pests or pathogens. Currently we have developed the Phytoseq panel that targets more than 30 genome regions in Phytophthoras and allows amplification of 6000 bp (nucleotides) in pools of 96 individuals. This powerful tool allows the user to obtain genome sequence data directly from environmental samples as well as pure cultures and assign unknown samples to a Phytophthora clade, species and even lineages (Figure 2).
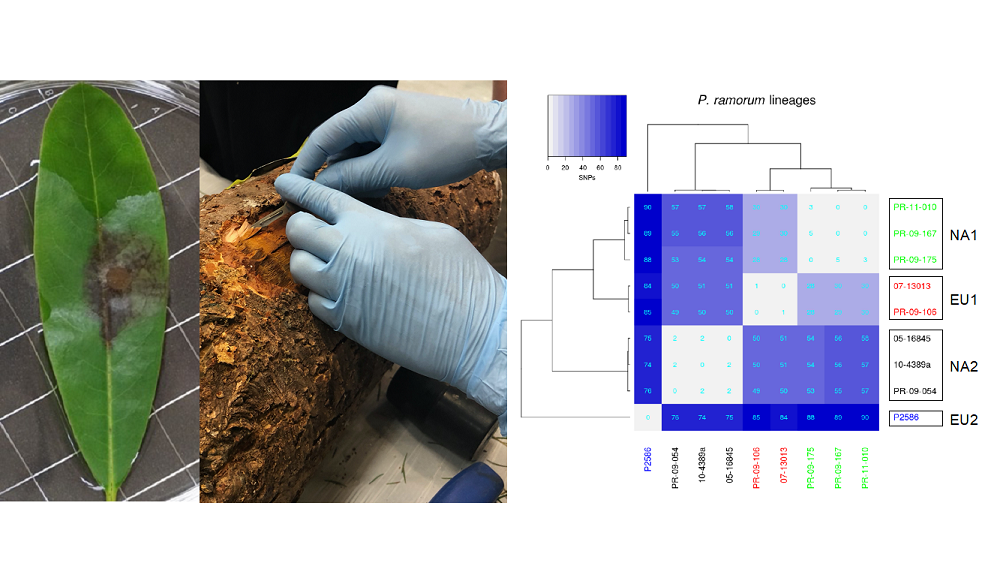
Figure 2. Amplification and sequencing of targeted genome regions directly from infected plant tissues and accurate assignment to lineages of P. ramorum.

